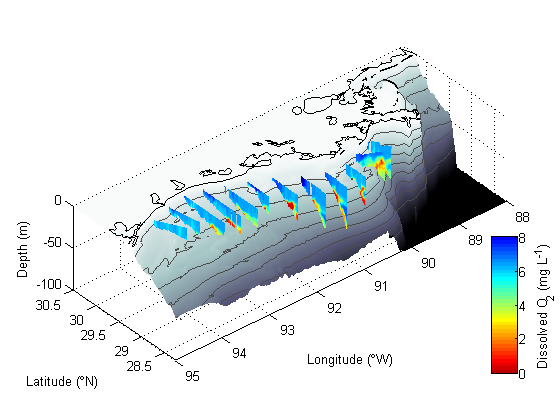
Making a map of nGOMEX region with curtain maps of oxygen
Contents
Example for MATLAB class
Part 0 - initializing the program
% Clear the workspace and close all open figure windows clear close all % Here I'm defining variable names, the limits for the color axis and the % string that we will use to label the colorbar. The variable name and % label are cells, the color axis is a 1x2 array. vars={'O2'}; cls=[0 8]; varn={'Dissolved O_2 (mg L^-^1)'};
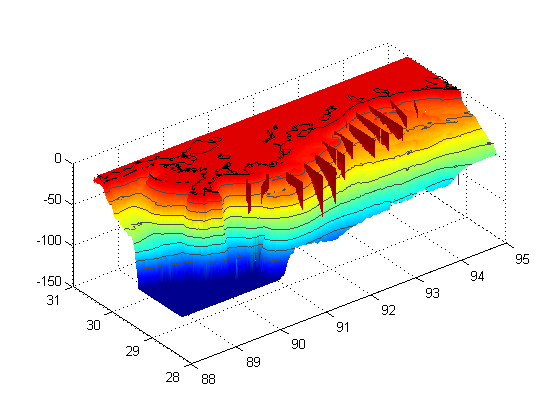
Part I - plot the bathymetry surface
The lines below load the bathymetry data I downloaded from NOAA, then adjust the values so that anything above the surface of the water is set to 0 and anything deeper than 100 m is set to 100 m. These are conventions purely for plotting.
load NGOMEX_seabathy_1min_NEW depth(depth>=0)=0; depth(depth<-100)=-100; % These lines make a surface plot with the bathymetry, set the 'edgecolor' % property of the surface to 'none', and set the aspect ratio of the plot % based on a Mercator projection. The depth is shifted by 1.5 m purely to % allow the contour lines (next part) p=surf(-X,Y,depth-1.5);shading interp;hold on set(p,'edgecolor','none') set(gca,'dataaspectratio',[1 cos(nanmean(nanmean(Y'))*pi/180) 60]); hold on
Part II - plot contour lines of certain isobaths
% Here we plot contour lines for certain isobaths, and shift these upward % by slightly more than the surface. This allows these lines to sit on top % of the surface plot so they are visible. The loop below the countour % call will set the colors of the contour lines by calling the individual % handles for each line and setting the colors. [c,ch]=contour3(-X,Y,depth-1,-80:10:-10,'k-'); for i=1:length(ch) set(ch(i),'color',[0.3 0.3 0.3]); end
Part III - loading and plotting the coastline
% Here we load the coastline file, truncate it to fit in our plot, and then % plot it as a dark black line. load NGOMEXcoast coast(coast(:,1)<-95 | coast(:,1)>-88 | ... coast(:,2)<28.25 |coast(:,2)>30.5 , :) =[]; plot(-coast(:,1),coast(:,2),'k-','linewidth',1);
Part IV - curtain maps of the oxygen data.
% First we are setting up the different transect line names as a cell array TRS={'B';'C';'D';'E';'Fa';'Fb';'G';'H';'J';'K';'Ia';'Ib';'W';'Q'}; % Here we loop through those transect line names... for i=1:length(TRS); % ... and load them sequentially fname=['RAPID_2010_',TRS{i},'.mat']; load(fname); % The line below is left over from a previous version of this program % that cycled through not only the transect lines, but also different % variables, to plot various variables for each transect. Basically it % changes the variable name to "V". eval(['V=',vars{1},';']); V=-V; % Here we careate a surface plot of the data and set shading to interp surf(-X,Y,-Z,V) shading interp % Keep.m is a function I got from the file exchange that clears all % variables except those specified. Helpful to reduce memory useage % http://www.mathworks.com/matlabcentral/fileexchange/181-keep keep depth i TRS coast p c ch vars cls varn end
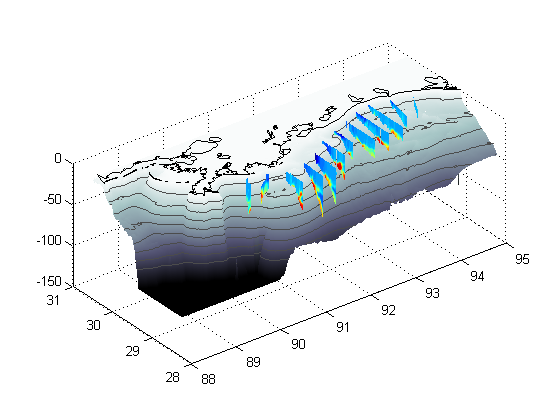
Part V - Set the colors
% Setcolormaps.m is from the web and allows the use of two colormaps within % one plot % http://code.google.com/p/guillaumemaze/wiki/matlab_routines_setcolormaps setcolormaps('bone','jet-1',-.000000001,512);
Part VI- the colorbar
% The lines below create a colorbar and then set various parameters for it % including the axis limits, the y-label, and the position (which includes % the size). h=colorbar('location','eastoutside'); set(h,'clim',[0 512],'ylim',cls) set(get(h,'ylabel'),'string',varn{1}) pos=get(h,'position'); pos(1)=pos(1).*1.07;pos(4)=pos(4).*0.4; set(h,'position',pos);
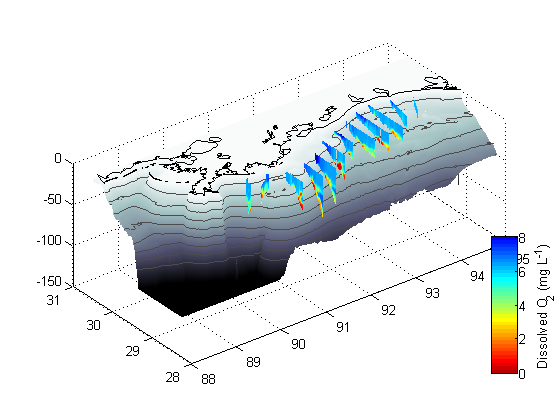
Part VII - make it pretty
% These lines simply add axis titles, set axis limits, and rotate the % figure to an aesthetically pleasing angle. set(gca,'xdir','reverse') xlabel('Longitude (\circW)'); ylabel('Latitude (\circN)'); zlabel('Depth (m)'); axis([88 95 28.25 30.5 -100 0]) view([-40 35]) shg